Key takeaways
- Highly sensitive gene and transcript detection with Evercode WT v3
- Consistently distinct cell clusters with version improvements
Sensitive gene and transcript detection in single-cell RNA sequencing (scRNA-seq) of human PBMCs is essential. It enables precise identification and characterization of various cell populations, including rare types, unveiling nuanced gene expression distinctions, and gaining insights into the dynamics of the immune system, responses to diseases, and potential therapeutic targets.
Here, cryopreserved PBMCs from 4 donors were purchased from a commercial vendor. Vials were thawed in a 37༠C water bath, transferred to a 50 mL centrifuge tube, diluted dropwise with warm FBS media, centrifuged, and washed with cold FBS. All samples had a viability >90% after thawing.
Samples were fixed with Evercode Cell Fixation v2 and Evercode Cell Fixation v3 kits and frozen at -80༠C.
Fixed samples were processed with the respective Evercode WT v2 and Evercode WT v3 kit. After barcoding, 57% of cells were retained for Evercode WT v3. For each kit, 4 sublibraries were loaded with 12,500 cells. All sublibraries were processed through the library prep workflow.
Whole transcriptome sublibraries were sequenced with an Illumina Novaseq 6000 on 1 lane of a Novaseq S4 flow cell.
After demultiplexing, sequencing data was processed with the Parse Biosciences data analysis pipeline v1.2.0. Results from all 4 sublibraries were analyzed from Evercode WT v3 for a total of 37,699 cells. Data was downsampled to an average of 40,000 reads/cell, integrated with Seurat, filtered to remove low quality cells, cell types classified with Azimuth, and annotations finalized manually. Median genes per cell was 2,447 at a read depth of 39,996 mean reads per cell. The analysis revealed all expected cell types in appropriate proportions, as shown on the UMAP below (Figure 1).
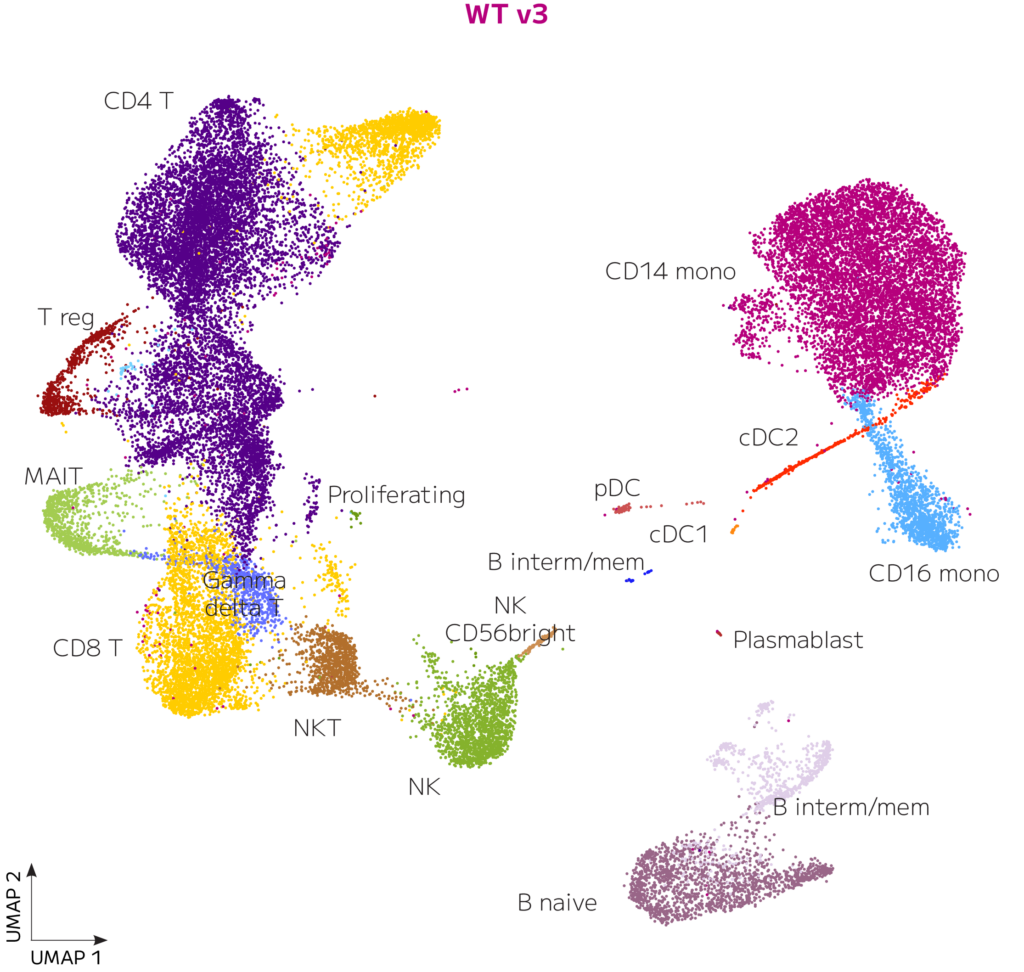
Figure 1. Healthy Donor Human PBMCs with Evercode WT v3. 37,699 PBMCs from 4 donors were processed with a single Evercode WT v3 kit. Sequencing data were processed with the Parse Biosciences pipeline, integrated with the WT v2 dataset, co-clustered, annotated, and visualized as a UMAP.
Next Steps
- Download the data files generated from the preparation and analysis of the human PBMC samples with Evercode WT v3 (below).
- Explore the dataset comparing the performance of Evercode WT v3 and Evercode WT v2 with these PBMC samples.
This dataset is licensed under the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) license. Non-commercial users are free to share and adapt the data by providing appropriate credit – Parse Biosciences citation guidelines are available here. If you’re interested in licensing the data for commercial purposes, visit our Dataset Commercial Licensing page.
Downloads
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.