TCR Sequencing of 1 Million Primary Human T Cells in a Single Experiment
Key Takeaways
- 1 million human T cells in a single experiment
- Sensitive clonotype detection at scale
Evercode TCR successfully uncovered nearly five hundred thousand unique beta chain clonotypes across 8 samples in a single experiment.
Pan T cells were isolated from 8 healthy human donors’ PBMCs and fixed using Evercode Cell Fixation Kit v2 to stabilize the cell structure and protect the RNA transcriptome from degradation. Fixed samples were stored at -80C until all samples were ready for further processing in a single experiment with the Evercode TCR Mega kit. Whole transcriptome and TCR specific libraries were sequenced on Illumina Novaseq 6000 S4 and S2 flow cells, respectively. The data was analyzed with Parse Biosciences Analysis Pipeline v1.0.4.
The assay demonstrated a high sensitivity, detecting TCR alpha chains (75% of cells) and beta chains (84% of cells) in primary human T cells. Median transcripts per cell were 1,420 while median genes per cell were 981 at a sequencing depth of 5,000 reads/cell. Clustering of cells using Seurat 4.0 resulted in all the expected major T cell subtypes in a single experiment, as demonstrated in the UMAP below.
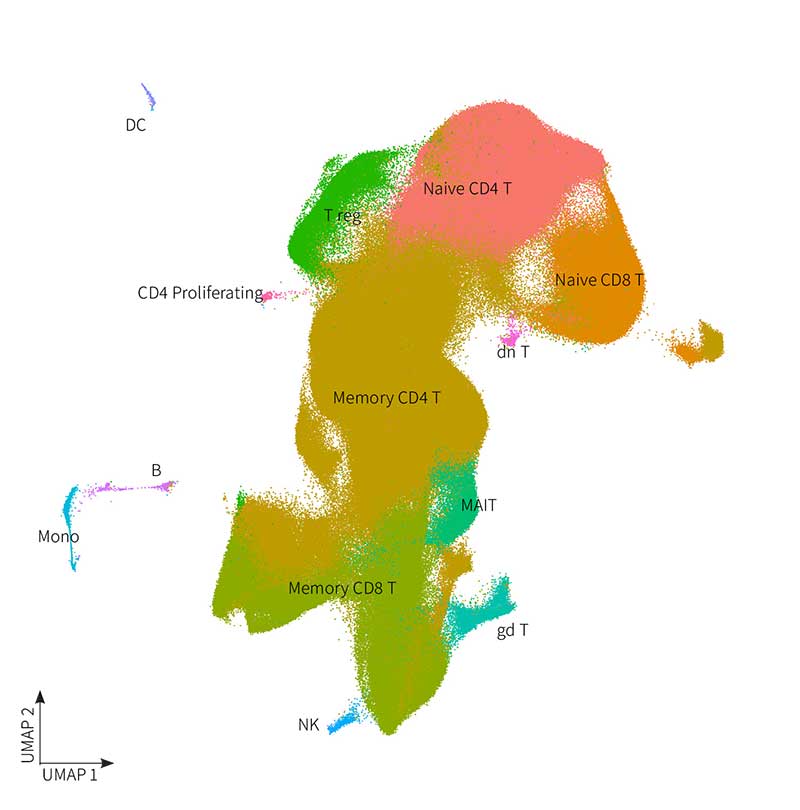
Figure 1: RNA clustering of T cells. Pan T cells were isolated from the PBMCs of 8 donors, fixed, and prepared together with an Evercode TCR Mega kit. All major T cell subtypes and a small percentage of infiltrating non-T cells were resolved in a single experiment.
Next Steps
- Download the data files generated from the preparation and analysis of the primary human T cells. These files include the experimental summary report for all data; clonotype frequency file, TCR barcode report file, and AIRR file for TCR data; the digital gene matrix file, the cell metadata file, and genes file for whole transcriptome data.
- Explore the activated T cells dataset generated from 80,000 cells.
- Review a more comprehensive description of Evercode TCR in the Evercode TCR Technical Brochure.
- Visit the Evercode TCR product page for Evercode TCR product highlights.
Downloads
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.