Key takeaways
- Sensitive gene detection with Evercode WT v3 kit at 100K scale
- Clustering reveals all expected PBMC cell types in appropriate proportions
Peripheral blood mononuclear cell samples (PBMCs) represent a heterogeneous population of cells, including lymphocytes (T cells, B cells, NK cells), monocytes, and dendritic cells. Analyzing PBMCs using scRNA-seq allows researchers to capture the diversity of immune cells present in the blood for understanding immune responses, diseases, and disorders.
Here, PBMCs from four healthy individuals were procured from a commercial supplier. The frozen samples were thawed in a 37°C water bath, transferred to a 50 mL centrifuge tube, diluted with warm FBS media, centrifuged, and washed with cold FBS. Following thawing, all four samples exhibited over 90% viability.
Subsequently, four fixation replicates of each donor were created with a population of 1 million cells each. These samples were then fixed using the Evercode Cell Fixation v3, 12 samples kit with 100 um plate strainers employing the mid-throughput plate-based workflow, and stored at -80°C.
After cell thawing, the four replicates of each donor were pooled and counted before proceeding with the barcoding process. Around 300,000 fixed cells were processed with Evercode WT v3, while two more aliquots were processed with Evercode WT Mini v3 and Evercode WT Mega v3.
A whole transcriptome sublibrary for Evercode WT v3 with 10,015 cells was sequenced on an Illumina® Nextseq™ 2000 along with 12,500 and 5,000 cell sublibraries from the other two kits.
Data processing was performed using the Parse Biosciences data analysis pipeline v1.2.0. Data from all sublibraries was downsampled to an average of 20,000 reads/cell, downsampled to the same number of cells, integrated with Seurat, filtered to remove low quality cells, cell types classified with Azimuth, and annotations finalized manually. Median genes per cell was 2,305 at a read depth of 31,984 mean reads per cell. The analysis revealed all expected cell types in appropriate proportions, as shown on the UMAP below.
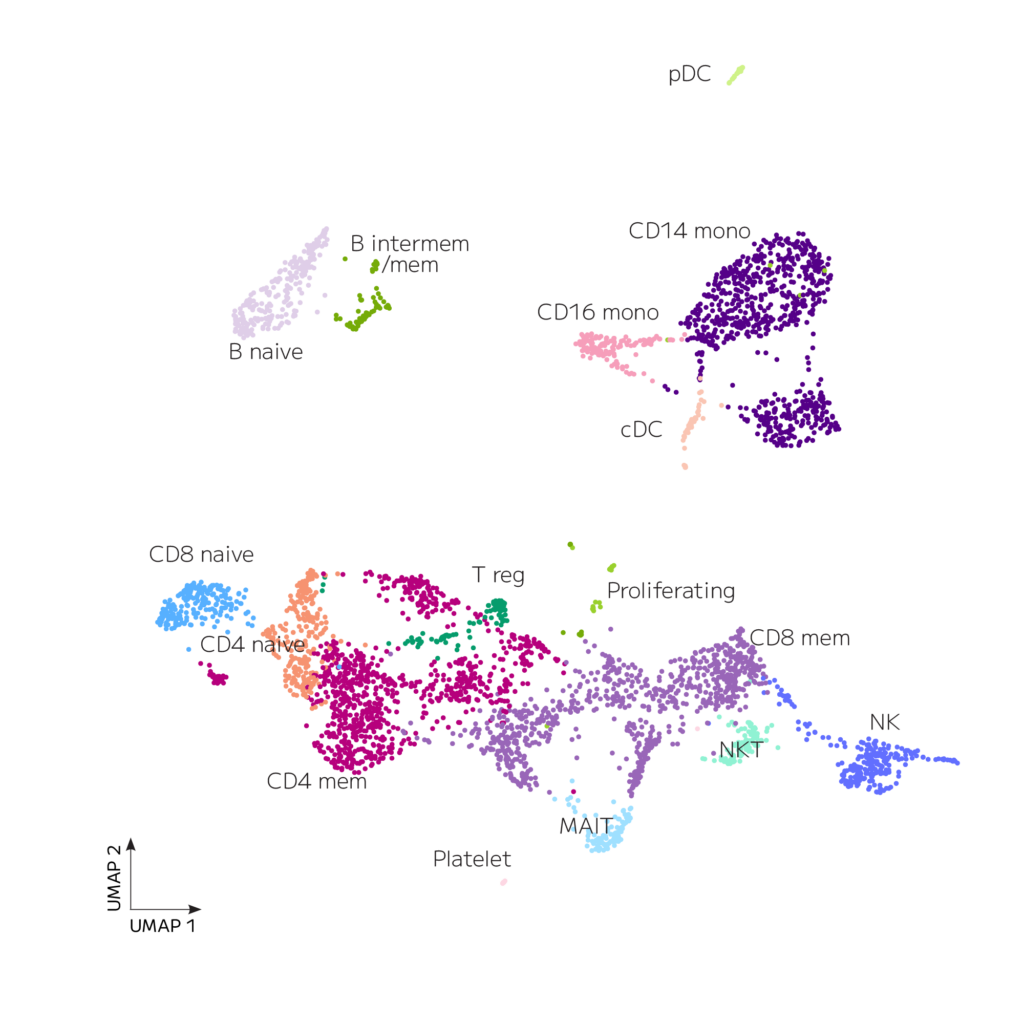
Figure 1. Human PBMCs with Evercode WT v3 kit. PBMCs isolated from 4 donors were processed with a single Evercode WT v3 kit to generate barcoded cells. After sequencing a whole transcriptome sublibrary with 12,500 cells, the data were processed with the Parse Biosciences pipeline, downsampled to 5,000 cells, integrated, co-clustered, annotated, and visualized as a UMAP.
Next Steps
- Download the data files generated from the preparation and analysis of the human PBMC samples with Evercode WT v3 (below).
- Explore the dataset comparing the performance of Evercode WT Mini v3, Evercode WT v3, and Evercode WT Mega v3 with these same samples.
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.