Key Takeaways
- Paired guide RNA detection and gene expression data with CRISPR Detect and Evercode Whole Transcriptome
- One guide RNA detected in >75% of cells
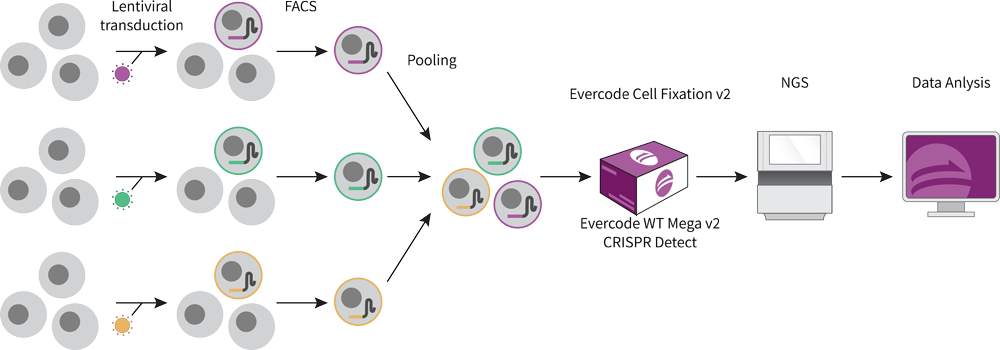
Figure 1: Sample Preparation Workflow for CRISPR Detect Performance Assessment.
To assess the performance of CRISPR Detect, cells were prepared to express just one guide RNA (sgRNA). Three guide RNAs were independently cloned into CROP-seq-Opti-eGFP and packaged into lentiviral particles. LN18 cells were separately transduced with the three lentiviral vectors at an MOI of 0.5. After 3 days of growth, GFP+ transduced cells were selected with FACS and pooled at an equal ratio. This experimental design examines the performance of the assay, by reducing background from biological variation (Figure 1).
After fixation with Evercode Cell Fixation v2, cells were processed with Evercode WT Mega v2 and CRISPR Detect. Whole transcriptome and CRISPR sgRNA libraries from one sublibrary were sequenced together on an Illumina Nextseq 550 with a High Output flow cell. The data was analyzed with the Parse Biosciences Analysis Pipeline.
At a sequencing depth of 3,000 reads per cell in the CRISPR Detect library, the library reached 98% sequencing saturation. The results reveal high overall sgRNA detection rates with 78% of cells having a single sgRNA as expected (Figure 2).
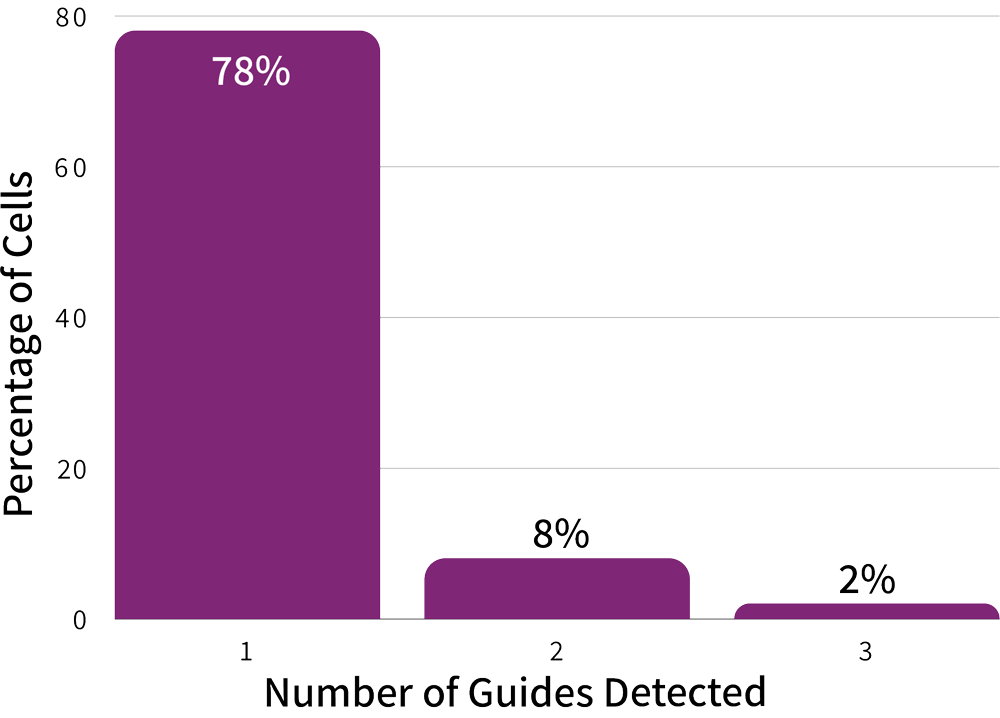
Figure 2: High sgRNA Detection Rate. 78% of cells were correctly assigned exactly one gRNA, while 8% and 2% of the cells exhibited two and three guides, respectively.
Next Steps
- Download the data files generated from the preparation and analysis of this sample. The datasets include the experimental summary reports, the digital gene expression matrix, guide assignment file, the gene files, and the cell metadata.
- Review a more comprehensive description of CRISPR Detect in the CRISPR Detect Product Sheet.
- Visit the CRISPR Detect product page for CRISPR Detect product highlights.
Downloads
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.