Uncovering Glioblastoma Heterogeneity in Solid Tumor and Tumor-derived in vitro Models
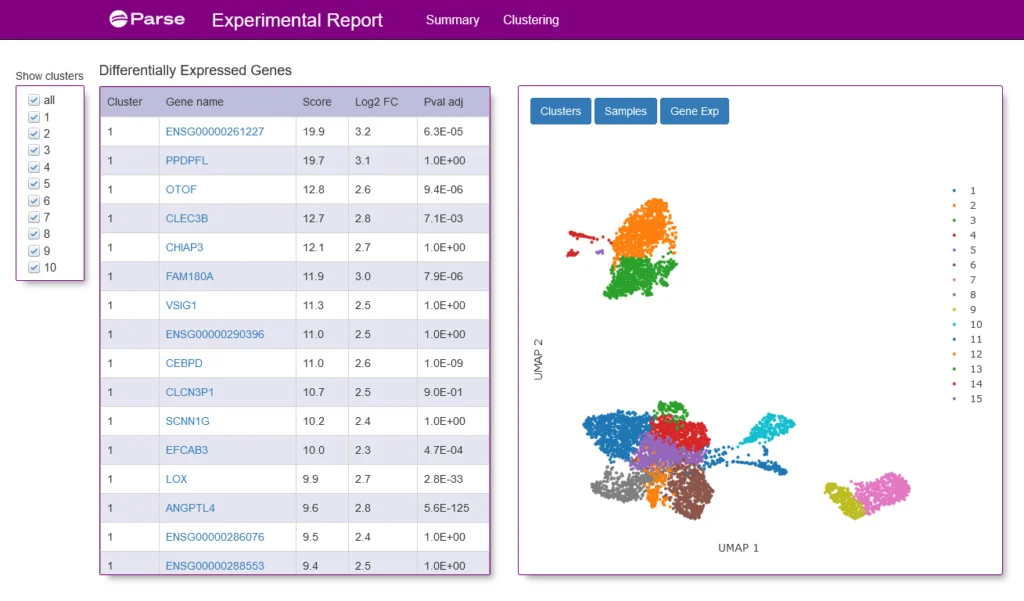
Glioblastoma remains the most common form of brain cancer, exhibiting enormous heterogeneity in the tumor and immune compartments, within both the same tumor and between patients. To characterize this heterogeneity within tumor samples and in vitro models, we utilized the Parse Evercode V2 assay to profile whole cells processed from a patient-derived neurosphere line, alongside nuclei isolated from a piece of flash-frozen glioblastoma tissue. Clustering of cells using Parse Analysis Pipeline revealed multiple distinct cell types from all samples, as demonstrated in the UMAP below.
Sample Table
| Donor | Material | Exposure | Processed | Name |
| 1 | Neurosphere | DMSO | Cells | Vehicle Cell |
| 1 | Neurosphere | TMZ | Cells | Treated Cell |
| 2 | Neurosphere | – | Nuclei | Sphere Nuclei |
| 2 | Frozen tissue | – | Nuclei | Tissue Nuclei |
* The plate heatmap of the all-well summary, wells 7-10 are empty, having been removed due to patient privacy.

"We obtained an unprecedented number of genes per cell."
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.