Evercode™ WT v2 vs. Chromium™ Next GEM Single Cell 3’ Kit v3.1 in Mouse Brain Nuclei

Key Tech Note Takeaways
- Head-to-head sensitivity comparison shows a marked contrast in gene detection.
- Cell type proportions are equivalently represented.
- Evaluation of differentially expressed genes reveals a disparity in the information attainable with each technology.
Unbiased Experimental Design
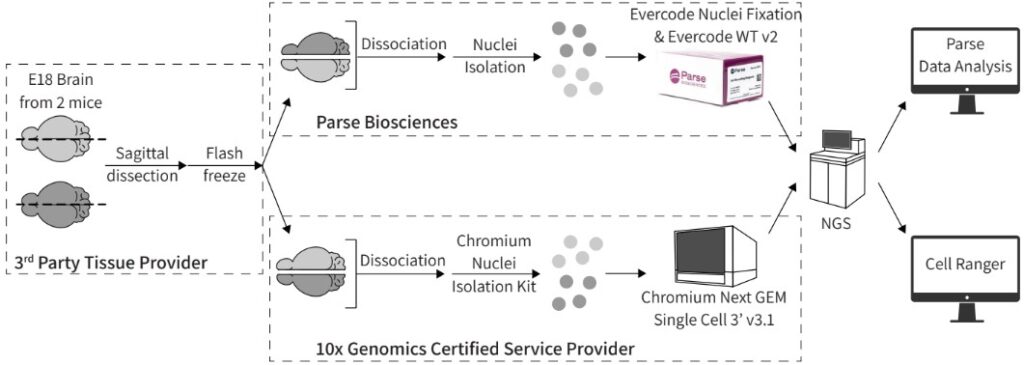
Two embryonic mouse brain samples were collected, sagittally dissected into 2 halves, and flash frozen by a third-party tissue vendor. Half of each brain was shipped to a 10x Genomics Certified Service Provider to isolate nuclei and create sequencing libraries. The other halves of each brain were processed by Parse Biosciences for nuclei isolation, fixation, and library preparation. Sequencing libraries from each technology were sequenced by a third-party. The sequencing data were analyzed by each manufacturer’s data analysis pipeline.
Downloads
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.