Key takeaways
- High gene detection in large adult hepatocytes
- Sensitive detection of proliferating hepatocytes and other subtypes
- High purity from Miltenyi liver perfusion kit demonstrated with Evercode
The liver is the largest organ in the body and is critical for numerous functions from metabolism to drug processing. The majority of liver cells are hepatocytes which are the key players for these essential functions.
Isolation and single cell RNA-Seq of whole adult hepatocyte cells using droplet-based methods are significantly challenging due to these cells’ large size and fragile nature.
Here, fresh liver tissue was collected from an adult mouse by Miltenyi Biotec, and semi-automated ex vivo liver perfusion was performed using gentleMACS™ Perfusion Technology, which runs on the well-established gentleMACS™ Octo Dissociator with Heaters (130-096-427). The liver was stored on ice in MACS Tissue Storage Solution (130-100-008) until hepatocytes were isolated following the protocol from the Liver Perfusion Kit (130-128-030), and the optional density-gradient purification step using Debris Removal Solution (130-109-3980) was included. The single cell suspension was fixed with Evercode Cell Fixation v2. Whole transcriptome libraries were generated with Evercode WT v2. For all hepatocyte isolation filtering steps, a 100 um MACS SmartStrainer (130-098-463) was used. A 70 um filter was used to filter cells immediately before fixation.
Whole transcriptome libraries were sequenced on an Illumina Nextseq 550 high-output flow cell, and data for the resulting 9,994 cells were processed with Parse Biosciences Analysis Pipeline v1.0.5. Median genes per cell was 1,765 at a read depth of 22,768 mean reads per cell. Clustering with Seurat v4.0 revealed that 99.7% of cells were hepatocytes with minimal endothelial contamination and a distinct population of proliferating hepatocytes.
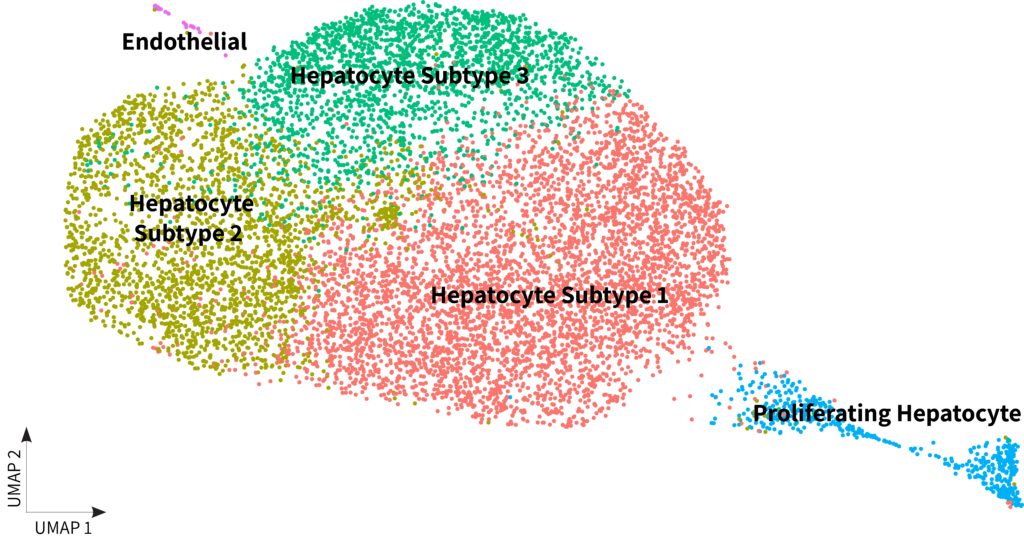
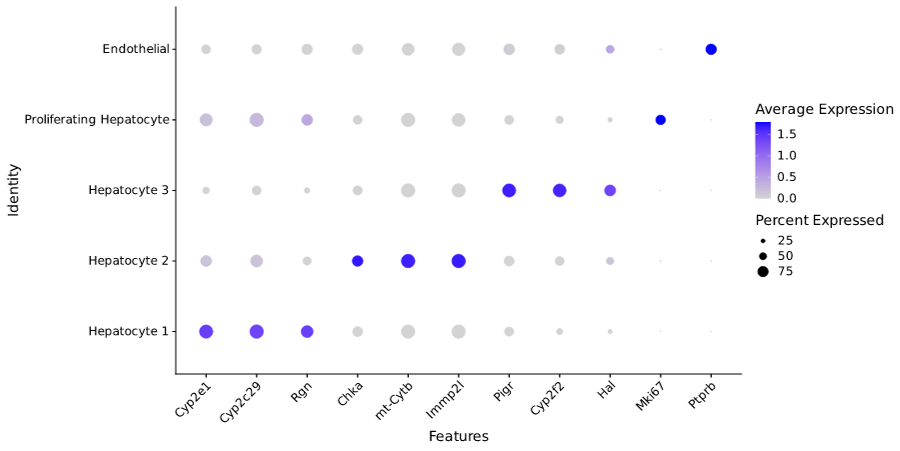
You can further explore the data and do your own analysis by downloading the raw data below.
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.