Performance of Evercode BCR in Mouse B Cells from 3 Different Mouse Strains
Key Takeaways
- Characterize the BCR repertoire of different mouse strains to uncover strain-specific immunoglobulin heavy and light chain characteristics
- Determine the BCR clonotype diversity and VDJ gene usage frequencies
Evercode BCR was used to profile over one hundred thousand mouse B cells from three different mouse strains – C57BL/6, PWK/PhJ, CAST/EiJ.
Pan B cells were isolated from the splenocytes of the mice and fixed using the Evercode Cell Fixation kit to stabilize the cell structure and protect the RNA transcriptome from degradation. Fixed samples were stored at -80°C until all samples were ready for further processing in a single experiment with the Evercode BCR, Mouse BCR + WT kit. Whole transcriptome and BCR specific libraries were sequenced on Illumina Novaseq X. The data was analyzed with Parse Biosciences Analysis Pipeline v1.4.1.
A)
B)
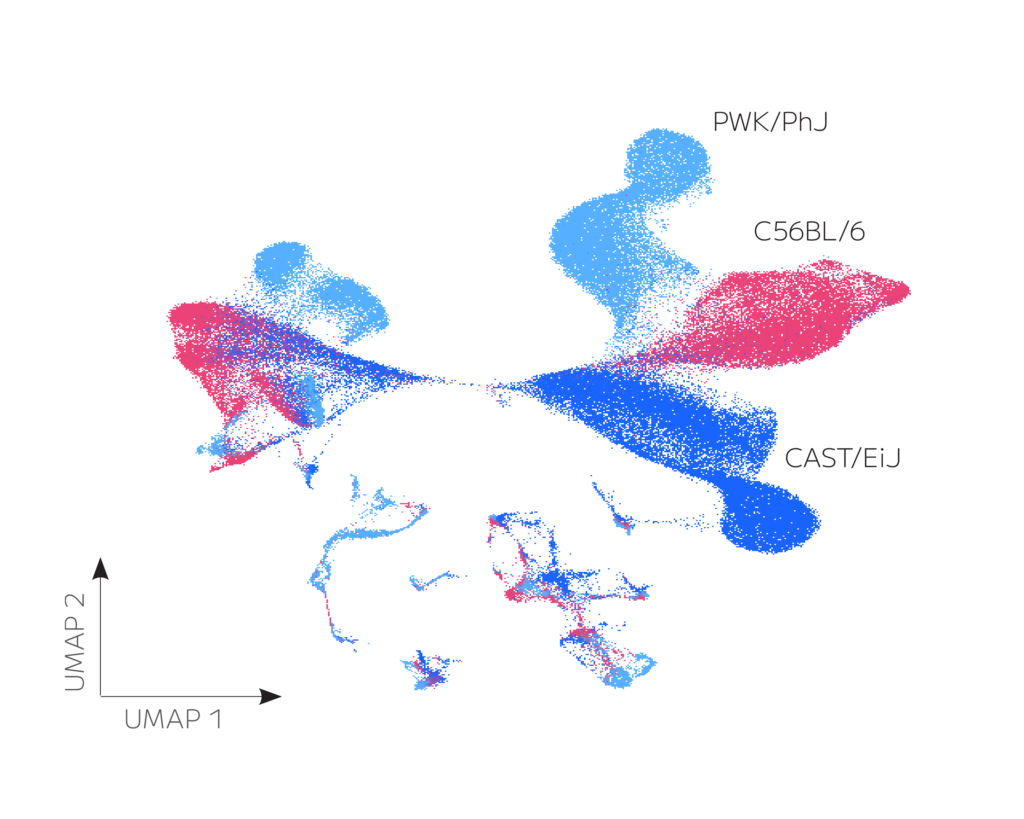
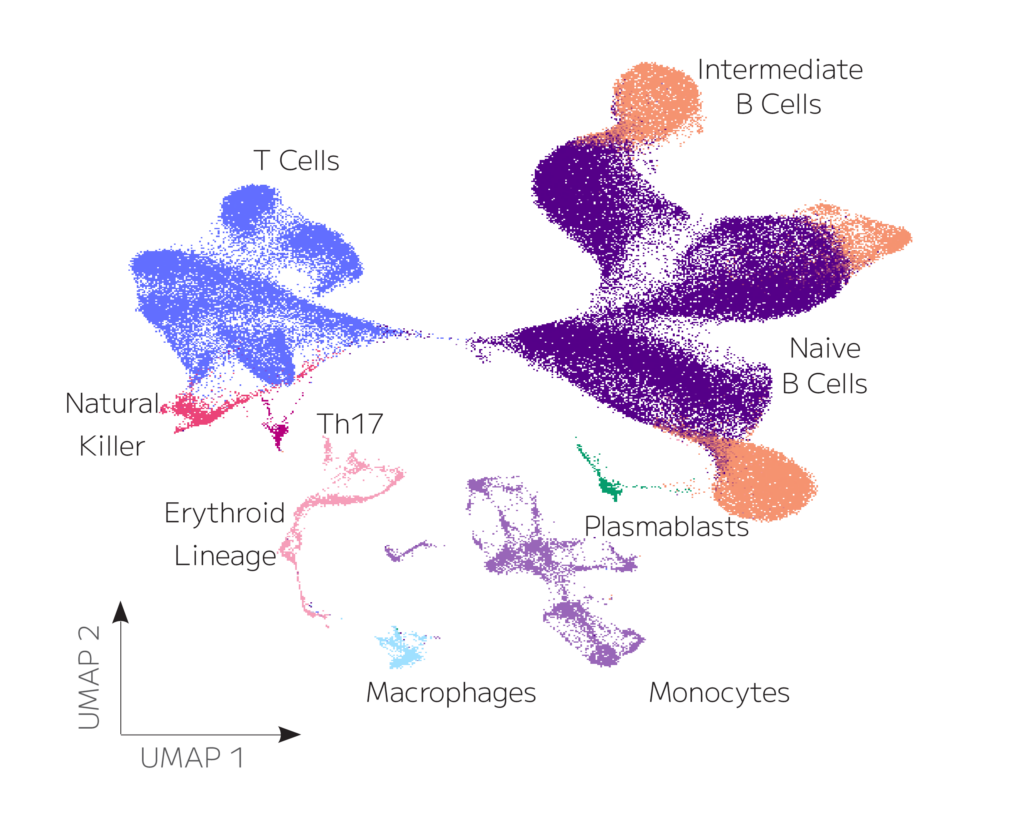
Figure 1: Transcriptomic clustering of 109,893 spleen cells pooled from three mouse strains reveals (A) cell- and (B) strain-specific identification and clustering of major B cell populations, including short-lived plasmablasts
Evercode Mouse BCR assay reveals strain-specific differential VDJ gene usage patterns that contribute to their unique and diverse BCR repertoire make-up, allowing for the selection of strains with suitable repertoire characteristics for the research study.
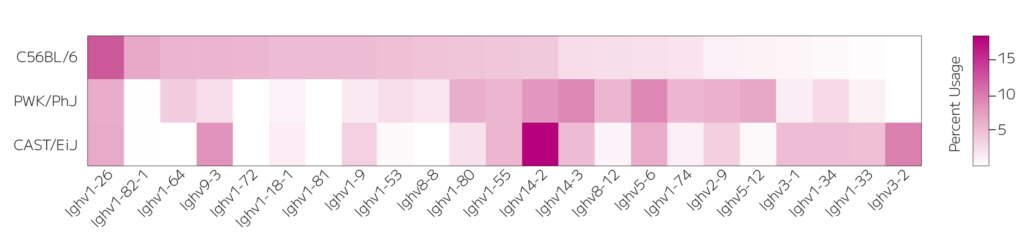
Figure 2: Heatmap of top V gene usage for three common laboratory mouse strains highlights distinct heavy chain V gene patterns, offering novel insights into their unique biology.
Next Steps
- Review a more comprehensive description in the Evercode BCR Technical Brochure.
- Visit the Evercode BCR product page for Evercode BCR product highlights.
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.