Evercode™ WT v2 vs. Chromium™ Next GEM Single Cell 3’ Kit v3.1 in Mouse Kidney Cells

Key Tech Note Takeaways
- Pronounced difference in gene sensitivity from the head-to-head comparison.
- Ambient RNA contamination significantly reduced when the cell is the reaction vessel.
- Higher resolution cell clustering attainable without the interferences of ambient RNA contamination.
Experimental Design
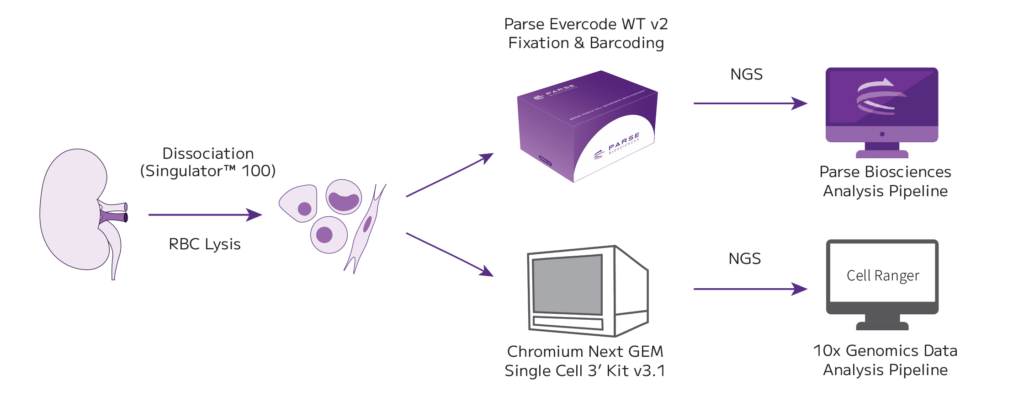
Mouse kidney tissue was dissociated into a single cell solution with a Singulator 100 (S2 Genomics). The samples were strained and red blood cells (RBCs) were lysed. The sample was split and half of the sample was prepared with the 10x Genomics Next GEM Single Cell 3’ Kit v3.1. The remaining half was fixed with Evercode Cell Fixation v2 and shipped for further processing with Evercode WT v2. Sequencing data were analyzed with each manufacturer’s respective analysis pipeline.
This dataset is licensed under the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) license. Non-commercial users are free to share and adapt the data by providing appropriate credit – Parse Biosciences citation guidelines are available here. If you’re interested in licensing the data for commercial purposes, visit our Dataset Commercial Licensing page.
Downloads
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.
