An Immunologist’s Experience with Human PBMCs with Evercode™ WT
Immunologist Dr. Howard Davidson from the University of Colorado recently profiled whole transcriptomes of peripheral blood mononuclear cells (PBMCs) from a healthy donor with Parse Biosciences Evercode technology. PBMCs are a heterogeneous population of primary immune cells where each unique cell type plays a distinct role in the immune system. Single cell RNA-seq of PBMCs is a powerful tool to help identify and characterize these unique cell types and their functional states.
Dr. Davidson collected and cryopreserved PBMCs from a healthy donor. After thawing, PBMCs were fixed using Evercode Cell Fixation v2 to maintain cell and RNA integrity. The fixed sample was shipped to Parse Biosciences for barcoding and library preparation with Evercode WT v2.
A whole transcriptome sublibrary for ~5,000 cells was sent back to Dr. Davidson and sequenced by the University of Colorado Anschutz core facility on an Illumina® Novaseq™ 6000. Initial data processing was done with Parse Biosciences Analysis Pipeline v1.0.3 and Seurat v4.3.0. Dr. Davidson’s analysis revealed the expected cell types, as shown on the UMAP below.
For more information on data analysis and comparing these findings with 10x Chromium Next GEM Single Cell 3’ Kit v3.1, see this Application Note:
A Researcher’s Head-to-Head Evaluation of Evercode™ vs. Chromium™ in Human PBMCs
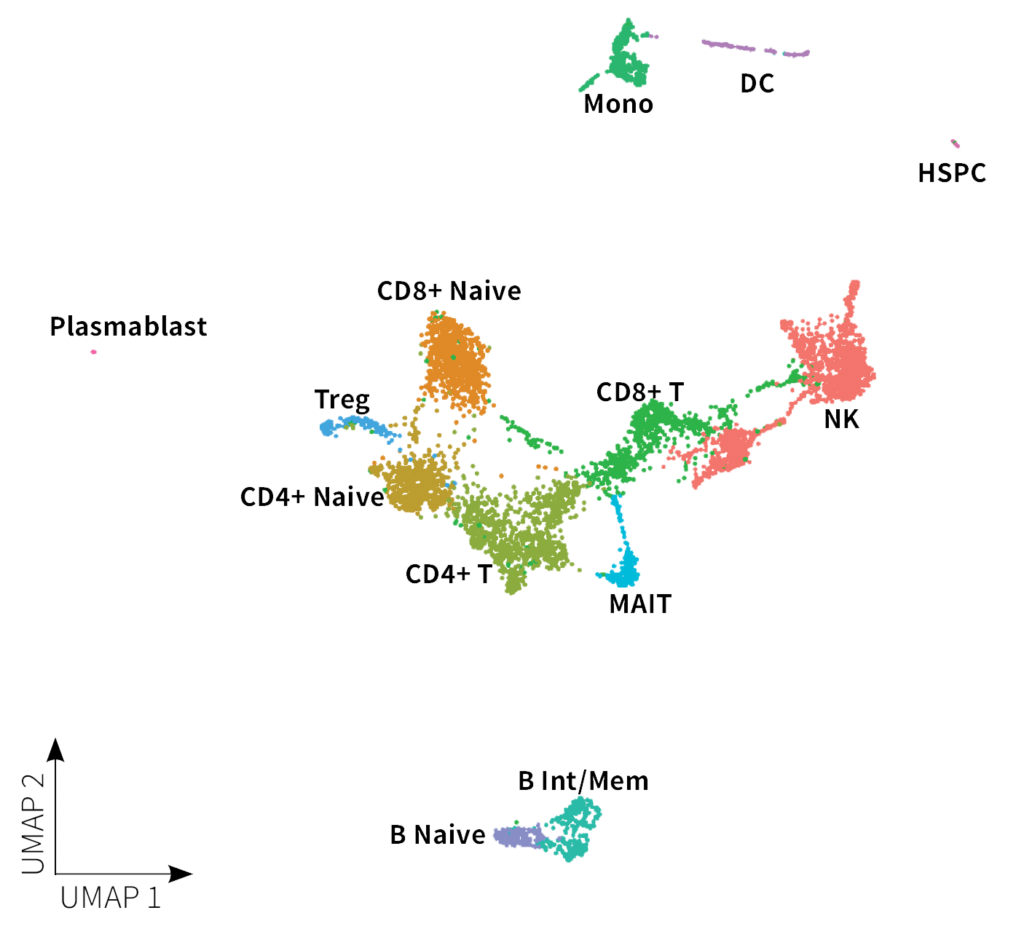
You can further explore the data and do your own analysis by downloading the raw data below.

"We looked at the results in many ways and remain impressed."
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.