Key Takeaways:
- Profiled 5 million nuclei from seven tissues in a single run using the Evercode WT Penta kit, avoiding batch effects and preserving sensitivity
- Mapped 211 distinct cell types, including rare and complex subtypes, and enabled analysis of sex-specific biology
- Demonstrates the power of large-scale single nucleus RNA-seq for building high-resolution, multi-tissue atlases
Experimental Design:
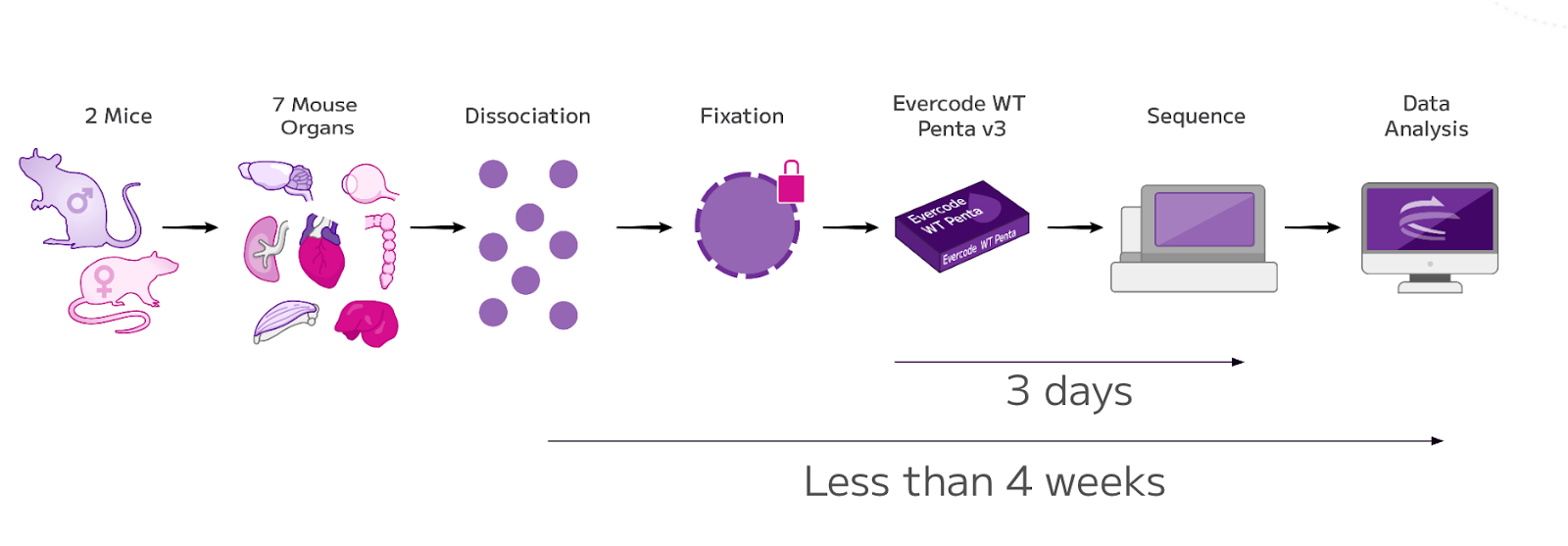
Figure 1: Experimental Overview. Nuclei isolated from seven flash-frozen mouse tissues were processed using Parse Biosciences’ Evercode Nuclei Fixation v3 and WT Penta kits, enabling the construction of a cross-tissue single cell transcriptomic atlas.
Tissues collected for the atlas included the liver, eye, heart, kidney, colon, quadriceps muscle, and brain, harvested from one male and one female mouse. These seven tissues were selected to represent a diverse range of organ systems. All samples were flash frozen immediately post-dissection and later processed for nuclei extraction.
Combinatorial barcoding was performed using the Evercode WT Penta kit, with asymmetric well loading to match sample complexity, ensuring appropriate representation and sensitivity across all tissues. A total of 32 sublibraries were generated, with one sublibrary sequenced initially to assess average reads per nucleus prior to full sequencing of the entire dataset.
Sequencing was conducted using a NovaSeq X, 100-cycle 25B flow cell, achieving high gene coverage with minimal saturation.
Results:
The final dataset comprises 5,011,382 nuclei spanning seven tissues collected from male and female mice (Figure 2). To generate the combined 5 million UMAP, nuclei were filtered using consistent thresholds across tissues: cells with fewer than 150 genes or more than 6,000 genes detected were removed, along with those exhibiting >30% mitochondrial content. Genes expressed in fewer than 10 cells were also excluded. Clustering and annotation reveal a rich landscape of cell types and states across major organ systems, enabling both cross-tissue comparisons and fine-grained exploration of individual tissues. This includes detection of rare populations, analysis of sex-specific biology, and identification of transcriptionally distinct subtypes within complex tissues.
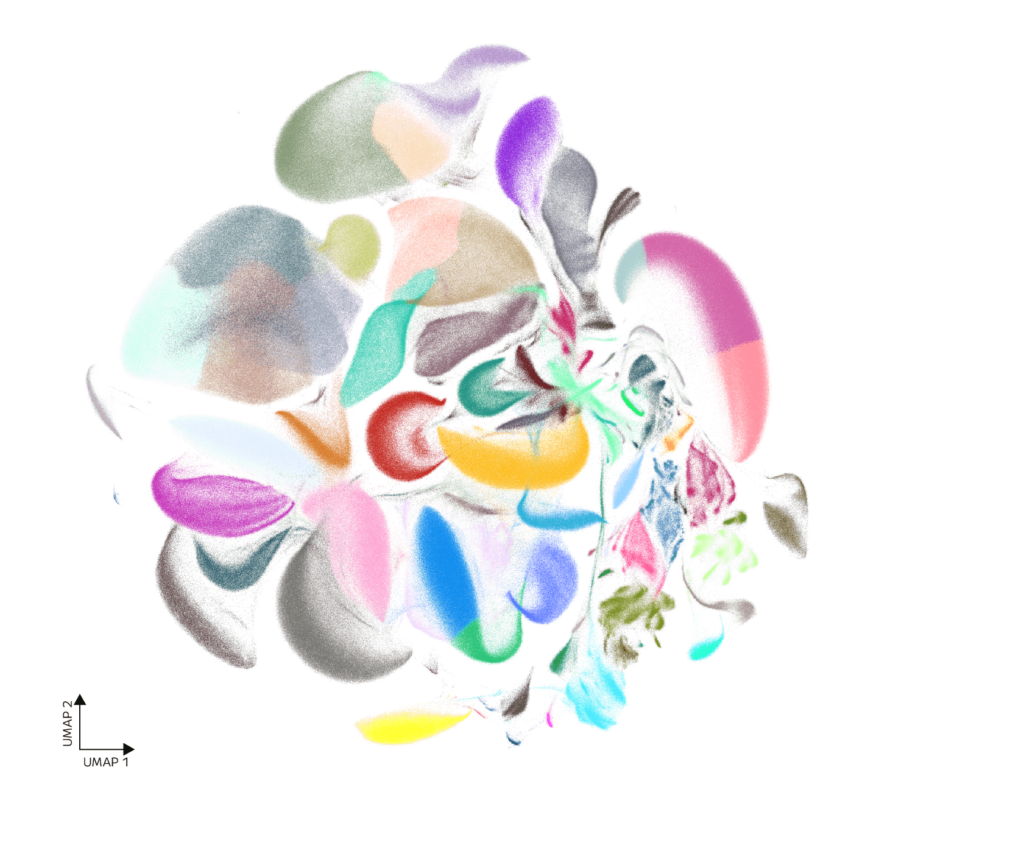
Figure 2: UMAP of 5 Million Mouse Nuclei.Distinct tissue-specific and shared transcriptional programs are resolved across cell populations.
Overall Experiment Metrics
- Total barcoded nuclei: 5,011,382 million
- Total annotated cell types: 211
- Median reads per nucleus: 9,741
- Sequencing saturation: 0.345
- Median transcripts per nucleus: 1,238
Importantly, 72% of all cell types had fewer than 10,000 cells, and 30% had fewer than 1,000 cells, underscoring the ability to capture rare and low-abundance populations. Despite this diversity, >95% of genes in the mouse reference genome were detected at a sequencing depth of 10,000 reads per cell, confirming excellent transcript coverage across the atlas (Table 1).
| Tissue | Protein-coding Genes | Non-coding Genes |
|---|---|---|
| Brain | 21,311 | 24,276 |
| Colon | 21,308 | 24,353 |
| Eye | 21,457 | 26,432 |
| Heart | 21,451 | 26,372 |
| Kidney | 21,404 | 25,781 |
| Liver | 21,460 | 26,283 |
| Quadriceps | 20,804 | 19,468 |
Table 1. Gene detection across tissues in the 5 million mouse nuclei atlas. Number of protein-coding and non-coding genes detected across all seven tissues in the 5 million mouse nuclei atlas. Gene detection was consistently high across tissues. This level of sensitivity, achieved across transcriptionally complex and diverse tissues, demonstrates the robustness of the Evercode WT Penta kit in delivering comprehensive transcriptome coverage at scale.
The eye dataset—a benchmark for transcriptomic diversity and rare population detection—demonstrated the resolving power of the Evercode Penta kit.
Clustering of 1 million nuclei from murine eye tissue revealed >100 distinct cell types, including keratinocytes, lens fiber cells, melanocytes, astrocytes, microglia, Müller glia, and retinal endothelial cells (Figure 3).
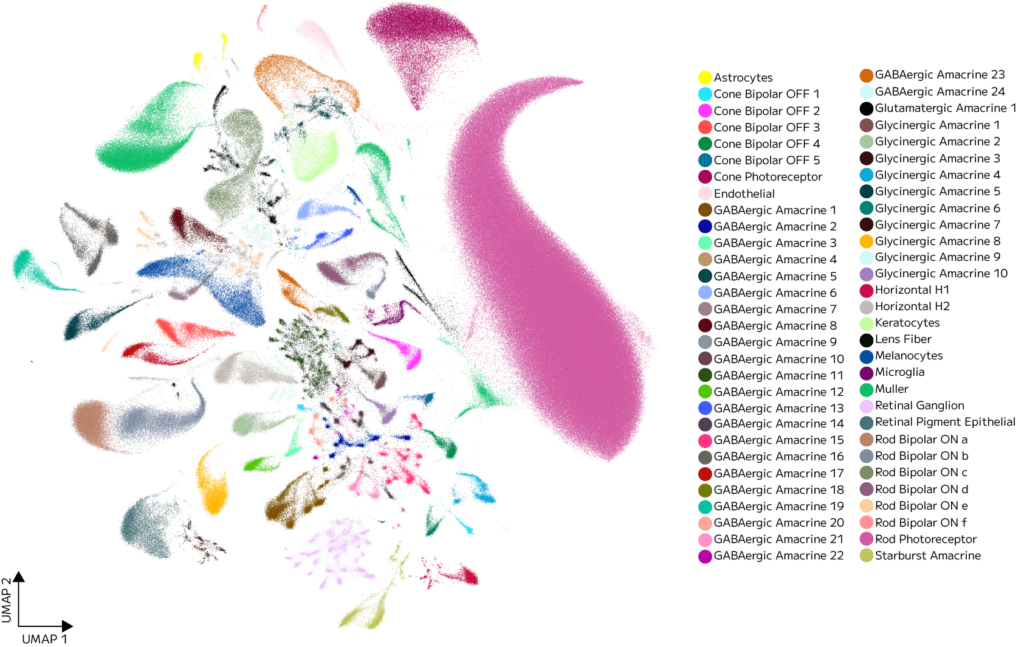
Figure 3. UMAP of the eye single nuclei atlas. Unsupervised clustering of the full dataset initially identifies 60 transcriptionally distinct cell type clusters, capturing the broad cellular diversity across the eye.
Focused sub-clustering of the retinal ganglion cell (RGC) population resolved 46 validated RGC subtypes, each with distinct expression signatures (Figure 4).
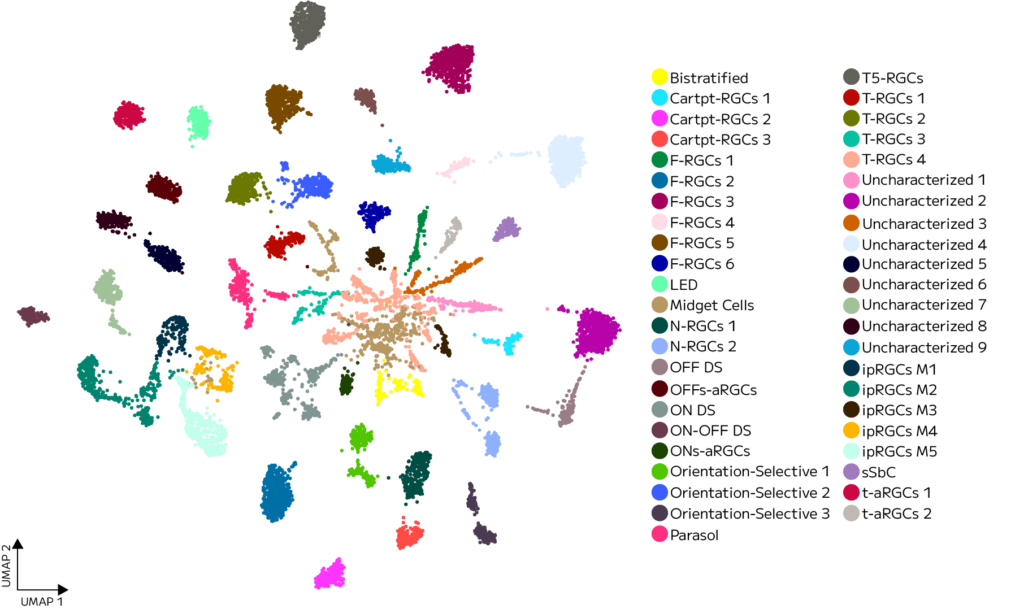
Figure 4: High-resolution UMAP of the retinal ganglion cell compartment. Sub-clustering of a single cluster reveals 46 transcriptionally distinct, marker-validated RGC subtypes.
Beyond RGCs, other populations show a clustering pattern visually consistent with further subtype diversity similar to the RGC cluster. These clusters are fertile ground for additional exploration and annotation by researchers using this dataset.
Why This Dataset Matters:
This dataset demonstrates the power of large-scale single cell for generating multi-tissue atlases. It offers:
- A reference-quality mouse nuclei atlas for studying organ-specific and shared expression patterns
- Insights into sex-based differences in gene expression
- Feasibility of generating species-agnostic atlases in your own lab under a month
Next Steps:
- Watch Evercode WT Penta Launch Webinar
- Explore the data using Trailmaker
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.