Single Cell Data Analysis Made Easy with Trailmaker
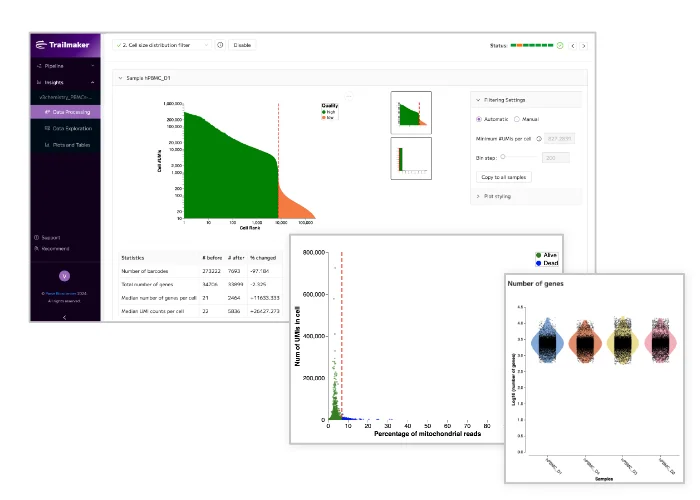
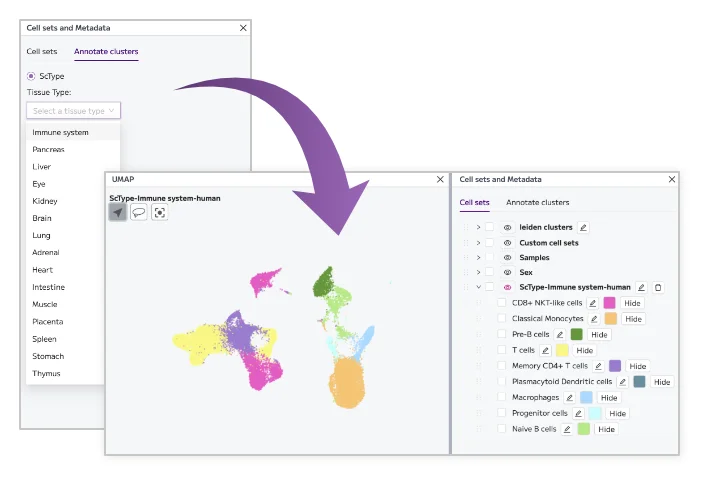
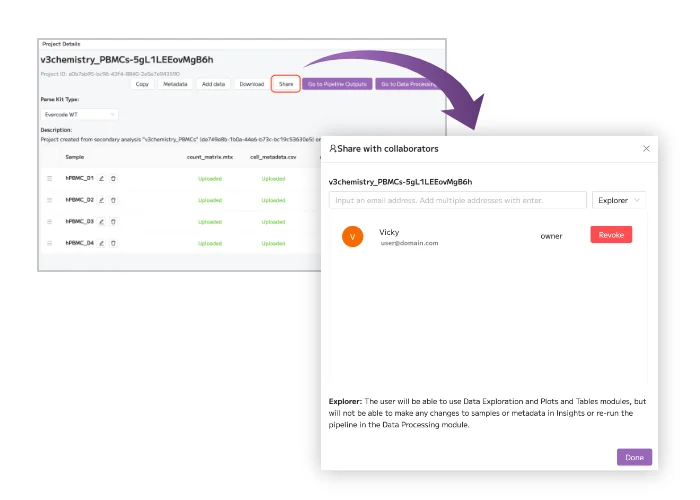
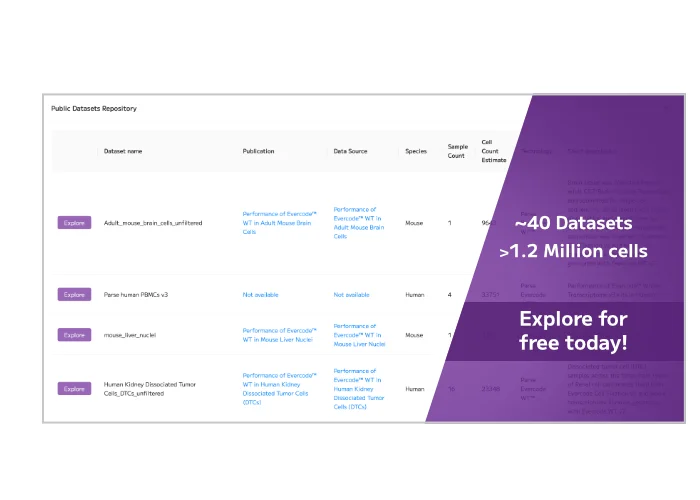
Unlock the power of single cell RNA-sequencing (scRNA-seq) with Trailmaker. Our user-friendly platform simplifies scRNA-seq data analysis, making it accessible and transparent for researchers. Quickly process, integrate, and explore Evercode Whole Transcriptome data, create publication-ready figures, and share your findings with ease.
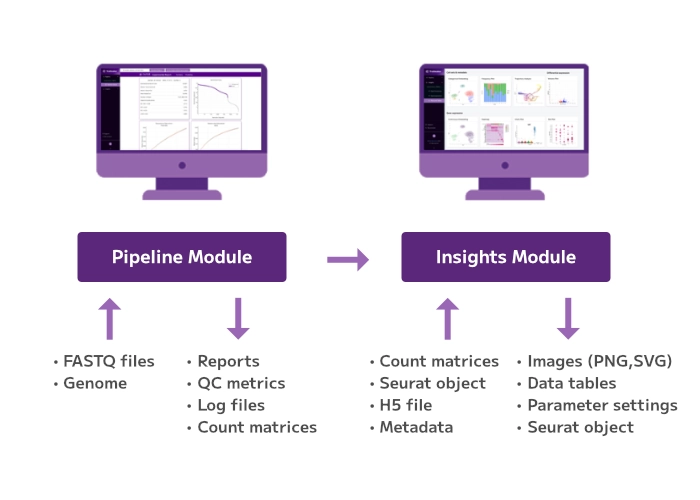
From Samples to Biological Insights
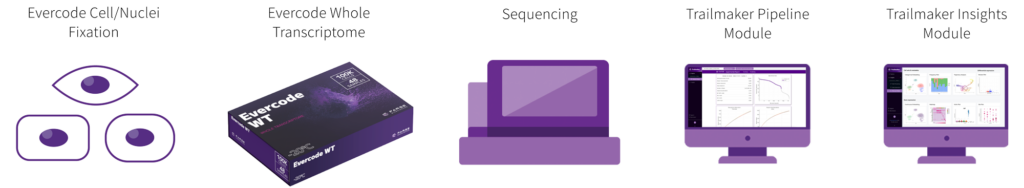
Our end-to-end workflow guides you from samples to discoveries, offering flexible entry, stopping, and exit points throughout the entire process, from sample preparation to data analysis.
Mastering Single Cell RNA-seq Data Analysis
This free course is designed specifically for biologists and accessible to all, requiring no coding experience. With a self-paced structure, comprehensive support, and guided learning through Trailmaker, it ensures an efficient and seamless experience.
 41 Lessons
41 Lessons 9 Hours of Video Content
9 Hours of Video Content Downloadable PDF Material
Downloadable PDF Material
Trailmaker Webinars On-Demand
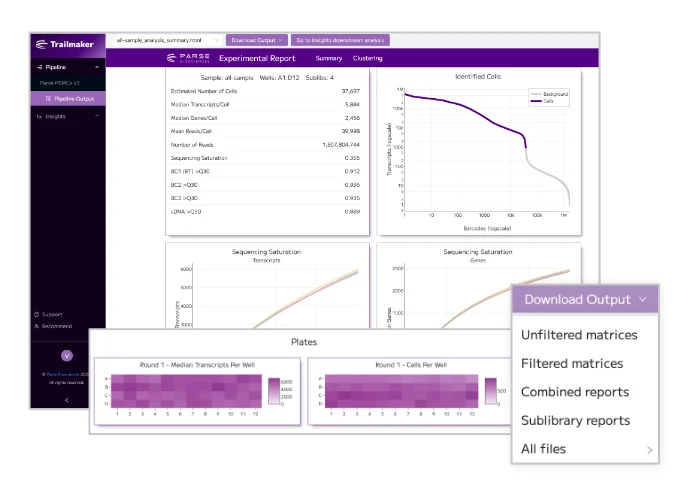
View our Trailmaker launch webinar and data analysis educational webinars on-demand. Our speakers will take you on a guided walk through of single cell RNA-seq data analysis from FASTQ files to publication-ready figures
Data Analysis: Resources and support
Download Trailmaker Product Sheet
Learn more about Parse Biosciences’ cloud analysis tools.

Looking for the original Parse Pipeline?
Parse Biosciences provides a pipeline that can be run on a Linux operating system,
either locally or on a server. The pipeline remains available for
customers to download via our support suite.