Uncovering Subtle and Divergent Responses in a Drug Screen Using Single Cell Sequencing with Evercode™ WT Mega 384
Key Takeaways:
- One researcher completed an anti-diabetic 88-compound, four-timepoint single cell drug screen in just four days.
- Resolved both strong perturbations and more subtle shifts within cell clusters that are best detected with single cell resolution.
- Showed that DPP-4 inhibitors, acting upstream of GLP-1 signaling, can be distinguished by unique temporal and transcriptional signatures, while also revealing potential off-target effects.
Experimental Design:
HEK293 and NIH-3T3 cells were seeded at 10,000 cells per well and grown to ~20,000 cells per well before treatment. Cells were then exposed to 88 small molecules from an anti-diabetic drug panel covering diverse mechanisms of action, including insulin sensitizers, sulfonylureas, and DPP-4 inhibitors, alongside DMSO controls (Figure 1) . The panel was designed to include compounds with both well-defined pathways and emerging modes of action, enabling assessment of how single cell readouts can differentiate drug responses.
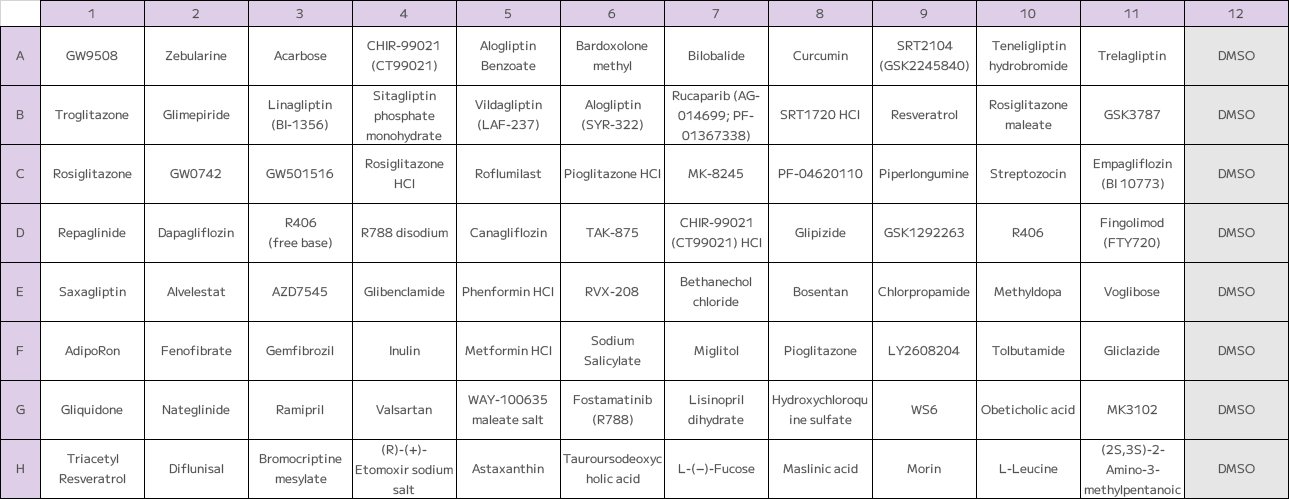
Figure 1: Plate layout of the 88-compound anti-diabetic screen.
Each well contained a single drug at 10 µM or a DMSO control, including diverse classes such as insulin sensitizers, sulfonylureas, and DPP-4 inhibitors.
Cells were fixed at 1, 3, 6, and 24 hours post-treatment using the Low Input Fixation workflow. This time-course design enabled capture of immediate, intermediate, and long-term transcriptional responses, while also revealing how these responses vary across different drug treatments.
All samples from different time points, cell lines, and treatments were processed in parallel, fixed, and run together downstream using the Evercode WT Mega 384 kit, minimizing batch effects. The entire workflow, from drug treatment through library preparation, was carried out by a single scientist in only four days.
Results:
Capturing Strong and Subtle Drug Affects
Single cell profiling resolved both dramatic and nuanced transcriptional responses. For example, Piperlongumine, an inducer of cell death, drove strong transcriptional shifts and cluster separation across time points (Figure 2). In contrast, Sitagliptin, a DPP-4 pathway inhibitor, produced a much more subtle shift within the existing cell type cluster (Figure 3).
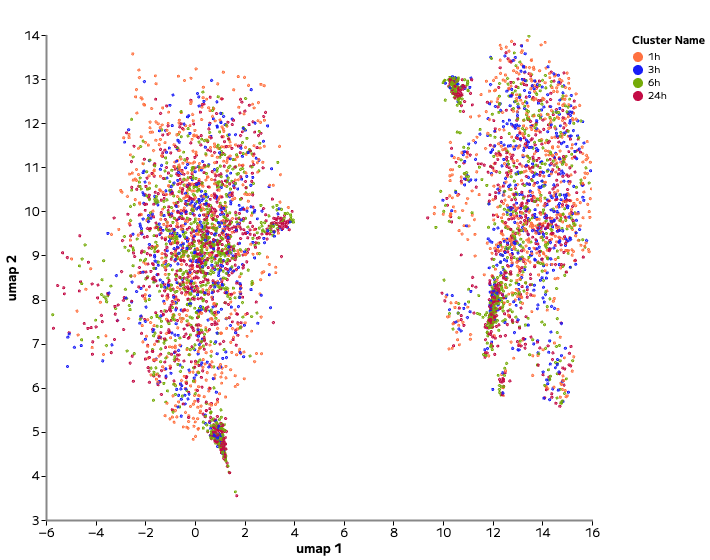
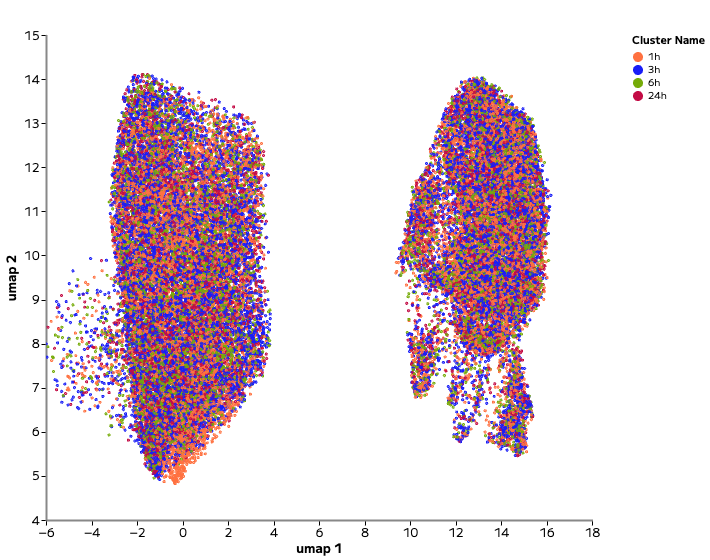
Figure 2: Drug with Strong Perturbation. UMAP showing transcriptional divergence following Piperlongumine treatment (top) versus control samples (bottom). Analysis and figure generation were performed in Trailmaker.


Figure 3: Drug with Subtle Perturbations. UMAP showing within-cluster shifts following Sitagliptin treatment (top) versus control samples (bottom). Analysis and figure generation were performed in Trailmaker.
Such subtle changes within the same cell line and drug treatment are best resolved with single cell resolution, where shifts within existing cell type clusters can be tracked over time. In bulk analysis, these changes might be dismissed as noise. However, by isolating subpopulations within the control cluster, single cell profiling makes it possible to identify the specific impact of drug treatment that would otherwise be missed. In some cases, these more subtle changes may reflect drugs that primarily modulate their on-target pathway with limited broader effects.
Divergent Responses Among DPP-4 Inhibitors
DPP-4 inhibitors displayed highly variable gene expression dynamics over time (Figure 4). Sitagliptin, for example, induced a dramatic transcriptional response, peaking at over 5,000 differentially expressed genes at 6 hours. In contrast, MK3102 produced more subtle effects, with an early peak at 1 hour that diminished quickly. Because the DPP-4 pathway lies upstream of GLP-1 signaling, these divergent responses may have broad downstream consequences. Volcano plots across time points (Figure 5) show how Sitagliptin’s impact evolves from 1h to 24h. By distinguishing genes consistently up-regulated across multiple time points, this analysis highlights that the single-cell approach is capturing genuine drug responses.
This dataset not only separates drugs targeting the same pathway by their diverse functional impact but also enables researchers to investigate potential off-target effects by examining specific genes and pathways perturbed beyond DPP-4 signaling.
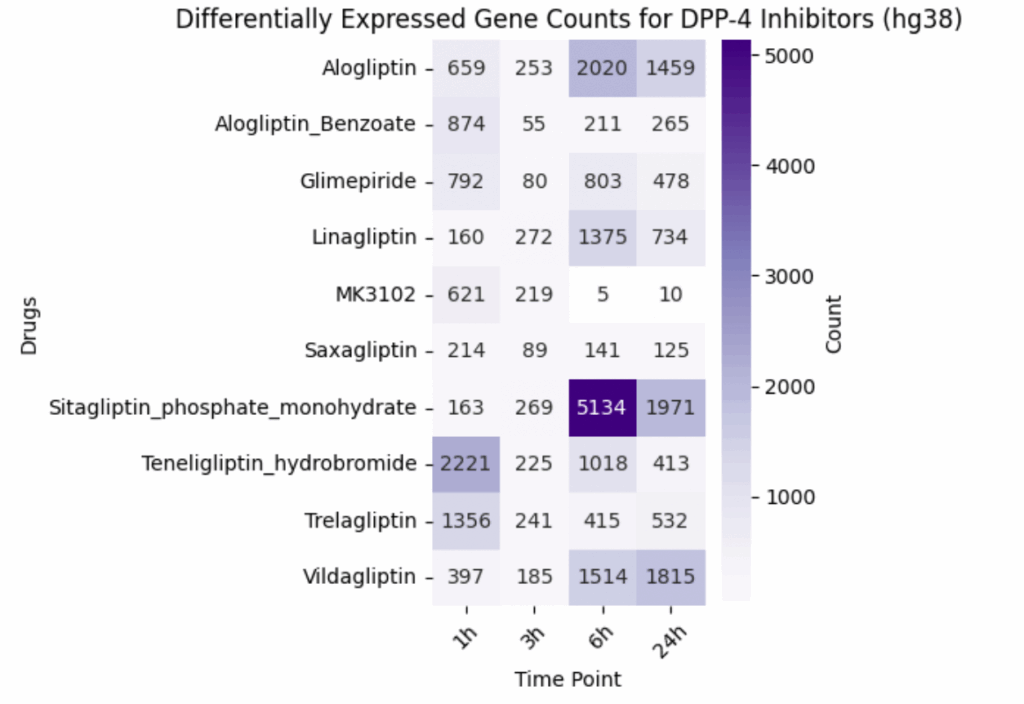
Figure 4: Variable Effects Among DPP-4 Inhibitors. Differentially expressed gene counts by compound and time point.
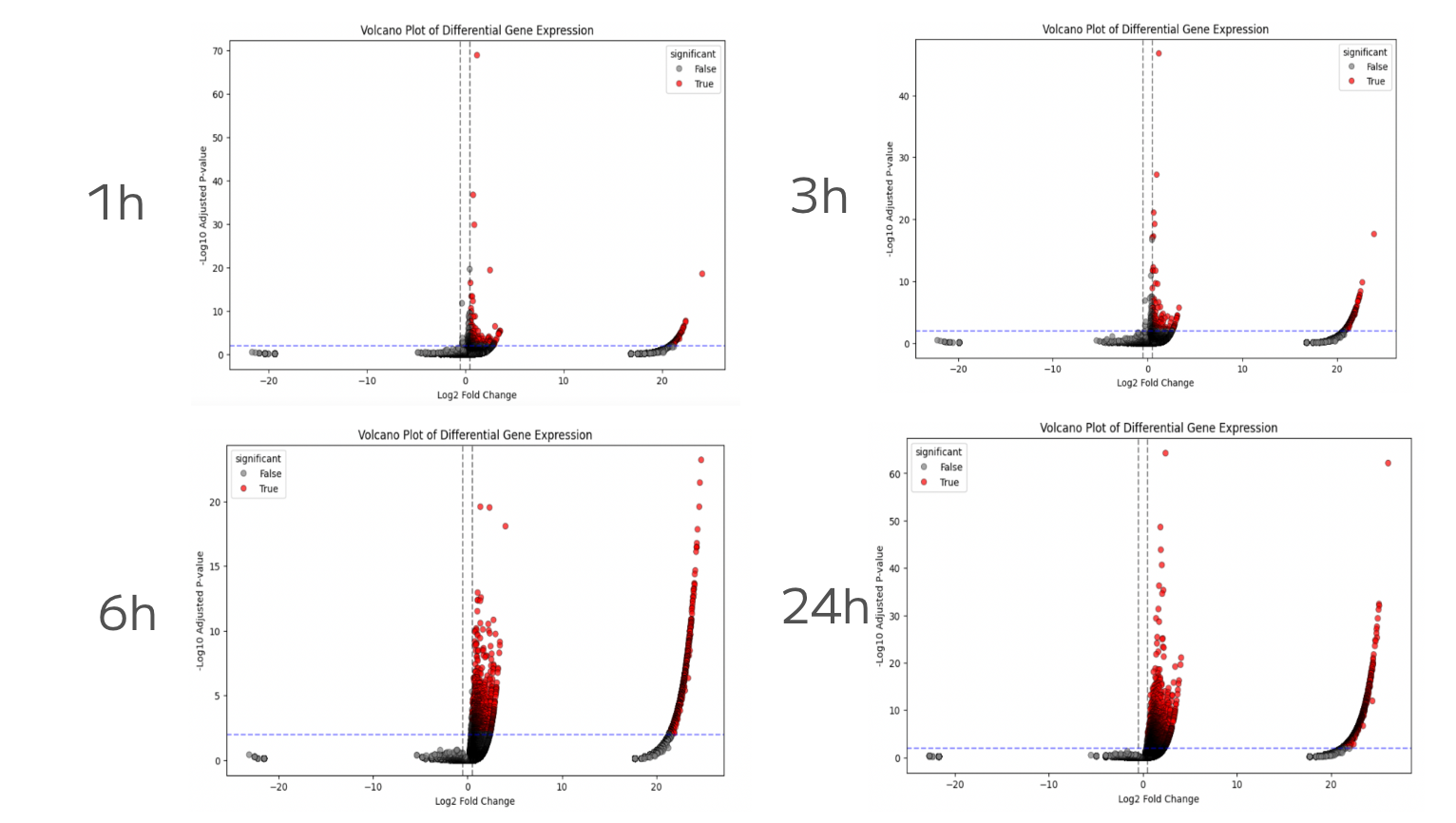
Figure 5: Temporal Dynamics of Sitagliptin Response. Volcano plots showing time-resolved differential gene expression from 1h to 24h. The two clusters illustrate the bimodality of gene changes: points on the far right represent strongly upregulated genes, while those closer to the center reflect more subtle but still statistically significant changes. Red dots indicate differentially expressed genes consistently observed across time points, whereas black dots represent noise.
This study shows how single-cell resolution transforms drug screening. With Evercode WT Mega 384, a single Parse scientist performed the entire drug screen and single-cell workflow through library preparation, 88 compounds across four time points, in just four days.
Single cell profiling captured both strong and subtle perturbations, including within-cluster shifts that are difficult to resolve with bulk approaches. The experiment also revealed that drugs with the same annotated mechanism of action, such as DPP-4 inhibitors upstream of GLP-1 signaling, exhibit dramatically different transcriptional responses over time. Together, these results highlight not only on-target effects but also provide a framework for studying off-target pathways and downstream consequences, positioning Evercode as a powerful tool for precise and comprehensive drug discovery.
This dataset is licensed under the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) license. Non-commercial users are free to share and adapt the data by providing appropriate credit – Parse Biosciences citation guidelines are available here. If you’re interested in licensing the data for commercial purposes, email us at .
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.