Evercode TCR Compared with Chromium V(D)J on Activated Human CAR T Cells
From a pool of activated T cells transduced with two different, FDA-approved CAR T-cell products, samples were processed with the leading two TCR single cell technologies. Results were compared for performance and data quality.
Key Takeaways
- 5X more cells assayed using the Evercode TCR assay drastically increased the value of the dataset
- Assays performance was comparable across all metrics
- The Chromium dataset had a pronounced fraction of ribosomal reads
Experimental Design
Human T cells were isolated from healthy PBMC donors, in vitro activated (anti-CD3, anti-CD28, IL2) and transduced with a lentivirus to introduce a CAR T-cell receptor. The constructs CD19 4-bb zeta, and CD19 CD28 zeta, target CD19 as therapy for B-cell lymphoma.
After ex vivo expansion, the cell cultures were prepared for the Parse Evercode TCR workflow or 10x Genomics Chromium Next GEM Single Cell 5′ Kit v2 with a Chromium Single Cell Human TCR Amplification Kit.
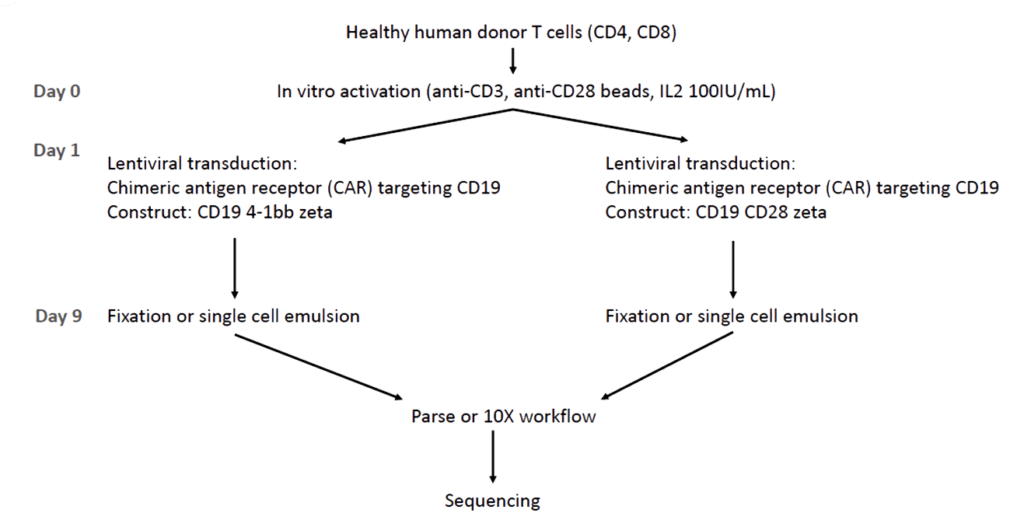
The libraries were then sequenced on an Illumina NextSeq 550 instrument, with FASTQ files processed using each technology’s specific pipeline. A single Parse sub-library was compared with a lane from the Chromium product. Sequencing results were compared after down-sampling to approximately 8k mean reads per cell for the whole transcriptome and 4k mean reads for the TCR data.
Whole Transcriptome Metrics
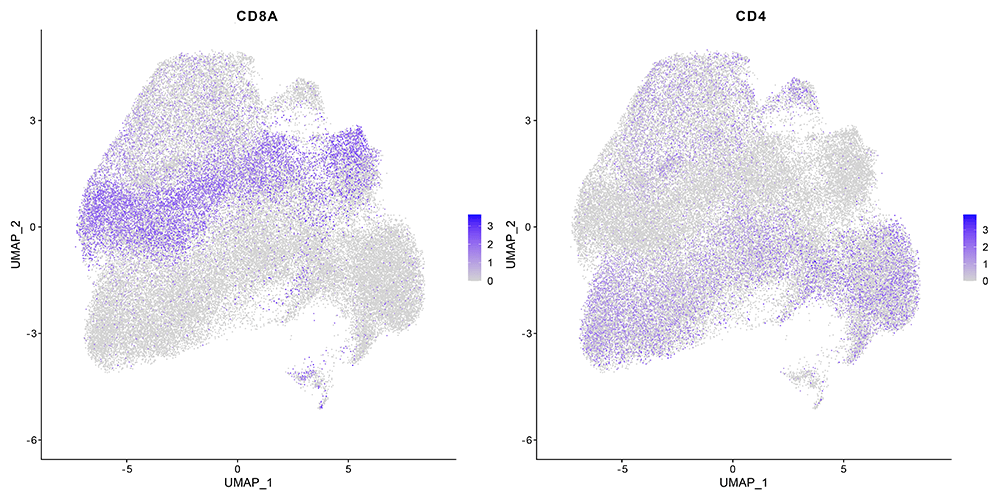
The Parse assay includes separate libraries and pipelines for the TCR and whole transcriptome data. Compared to the Chromium results, a highly similar fraction of cells with paired TCR chains and the number of genes per cell, were identified. All the major T cell subtypes are observed in both approaches.
| No. of Reads per Cell WT | No. of Reads per Cell TCR | Fraction Cells w/ a TCR | Fraction Cells w/ WT & Paired TCR | No. of Cells Identified | No. of Unique Clonotypes | |
| Evercode TCR | 7,781 | 3,962 | .989 | .870 | 50,012 | 42,493 |
| Chromium V(D)J | 7,826 | 3,992 | .935 | .908 | 10,276 | 9,180 |
Evercode TCR Identified Key Cell Type Markers Defining Broad T-Cell Subsets
Percent Ribosomal Proteins by Technology
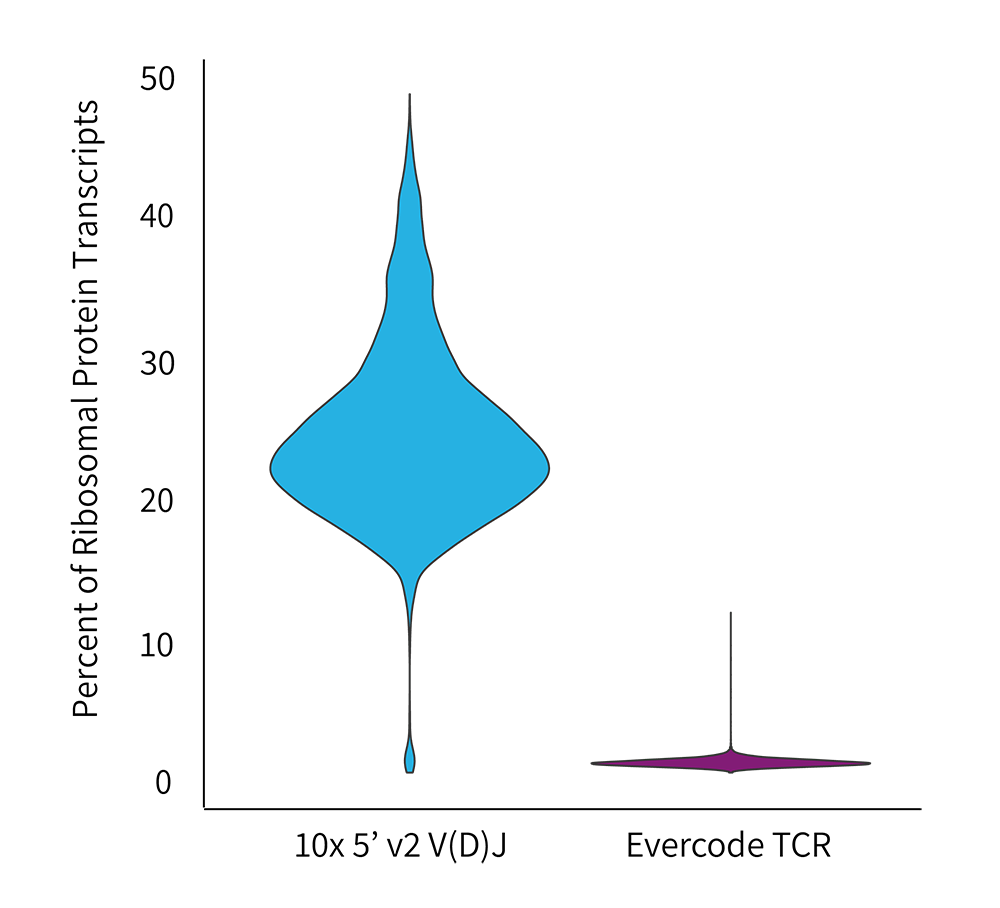
A significantly higher fraction of ribosomal proteins were observed in the Chromium dataset.
Total Unique TCRs Identified
Using the Evercode TCR assay, 5-fold more unique cells and clonotypes were obtained from a single sub-library than a single lane of the Chromium V(D)J assay. We note that a large TCR sample size is needed to observe rare clonal types and to track clones over time. A large sample size is important for observing sufficient numbers of T cells, given the highly variable proportion of T cells in different tissues, and effects of different treatment regimes. In addition, larger TCR samples provide a more accurate measure of T cell repertoire diversity (i.e., clonality) and V, D, and J gene usage.
With the Evercode TCR assay, samples are collected over time, fixing and running them all at once. This reduces batch effects and enables us to scale a research program.

"I compared technologies and I'm using Parse TCR for our next experiments"
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.