Key Takeaways
- Large, adult brain cells profiled
- Differentiation of the expected major brain cell types
- Comprehensive view into cellular states, dynamic processes, and alternative splicing
The brain, with its hundreds of billions of interconnected cells and trillions of connections, is the most complex organ and one that surpasses our comprehension. The afflictions that impair its function, such as Alzheimer’s disease, schizophrenia, Huntington’s disease, and various others, are some of the most agonizing and devastating conditions known to us. Single cell transcriptomic analysis offers valuable insights into the workings of this complex organ. Researchers often turn to single nuclei RNA sequencing (snRNA-Seq) as a solution to difficulties encountered with cell dissociation methods and the relatively larger sizes of cells in the brain, which are incompatible with microfluidic approaches. However, this approach leads to the loss of cytoplasmic transcripts. Evercode technology overcomes these limitations by enabling the collection of cytoplasmic transcripts even from large cells.
Here, fresh brain tissue was collected from an adult C57/BL6 mouse by Transnetyx and shipped for single cell gene expression experiments. After mechanical tissue dissociation and percoll clean-up (Mattei et al, 2020), the single cell suspension was fixed with Evercode Cell Fixation v2 and whole transcriptome libraries were generated with Evercode WT v2.
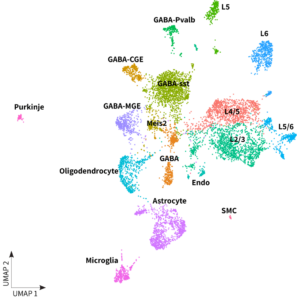
Whole transcriptome libraries were sequenced on an Illumina Nextseq 550 high-output flow cell, and data for the resulting 15,200 cells were processed with Parse Biosciences data analysis pipeline v1.0.5. Clustering of cells with Seurat v4.0 and additional analysis revealed the expected neuronal and non-neuronal cell types, as shown in the UMAP below. Additionally, it is worth noting the clear differentiation of Purkinje cells, one of the brain’s largest and unique neurons.
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.