Fresh lung tissue was collected from an adult CD-1 mouse and immediately processed. After isolation with the Singulator™ 100 (S2 Genomics™), nuclei were strained, centrifuged, and resuspended in Nuclei Storage Buffer. The single nuclei suspension was fixed with Evercode Nuclei Fixation v2 then whole transcriptome libraries were created with Evercode WT v2.
Libraries were sequenced on an Illumina Novaseq 6000, and data was processed with Parse Biosciences Analysis Pipeline v1.0.2. The resulting 14,097 nuclei were clustered with Seurat v4.0, manually annotated, and visualized as UMAPs. Our analysis revealed the expected cell types, as shown in the UMAP below.
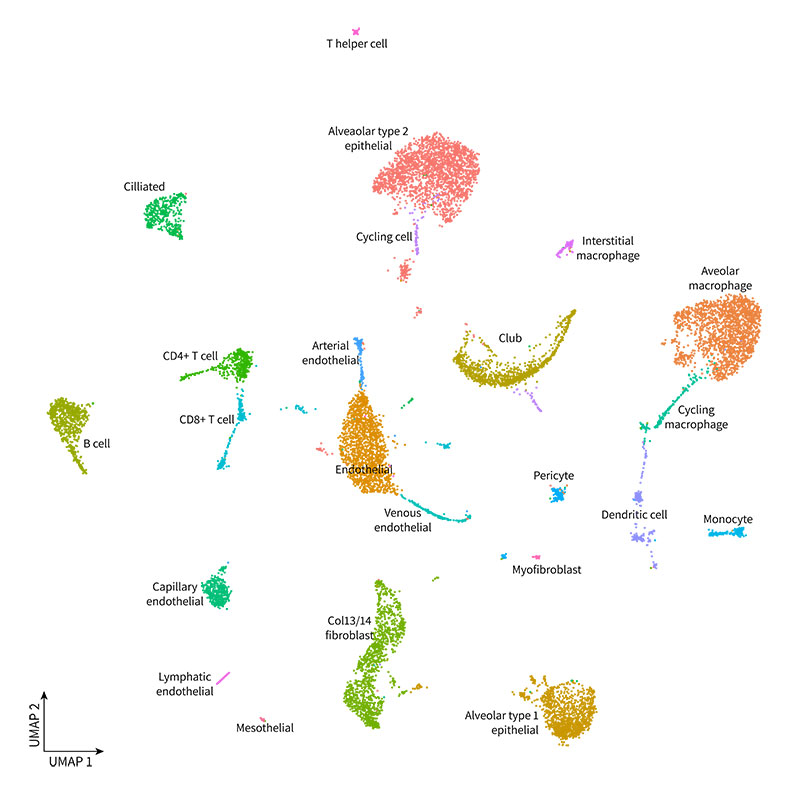
You can further explore the data and do your own analysis by downloading the raw data below.
We're your partners in single cell
Reach out for a quote or for help planning your next experiment.